Evaluation of explanation quality: saliency maps#
In this notebook, we are going to explore how we can use teex to compare image explanation methods.
1. Evaluating synthetic image data explanations#
In this section, we are going to 1. Generate image data with available g.t. explanations using the ‘seneca’ method. 2. Create and train a pytorch classifier that will learn to recognize the pattern in the images. 3. Generate explanations with some model agnostic methods and evaluate them w.r.t. the ground truth explanations.
[ ]:
%pip install teex
%pip install captum
%pip install torch==1.7.1+cu110 torchvision==0.8.2+cu110 -f https://download.pytorch.org/whl/torch_stable.html
[2]:
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
import torch
import torch.nn as nn
import torch.optim as optim
import torch.nn.functional as F
import PIL
import pickle
from sklearn.model_selection import train_test_split
from sklearn.metrics import accuracy_score, f1_score
from math import floor
from captum.attr import GradientShap, IntegratedGradients, Occlusion, DeepLift, Lime, GuidedBackprop, GuidedGradCam
from teex.saliencyMap.data import SenecaSM
from teex.saliencyMap.eval import saliency_map_scores
from teex._utils._arrays import _minmax_normalize_array
[ ]:
from google.colab import drive
drive.mount('/content/drive')
Mounted at /content/drive
1.1. Generating the data#
[3]:
cellH, cellW = 4, 4
imH, imW = 32, 32
generator = SenecaSM(imageH=imH, imageW=imW,
cellH=cellH, cellW=cellW,
nSamples=5000, randomState=7)
X, y, exps = generator[:]
pattern = generator.pattern
[4]:
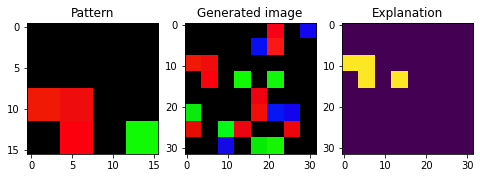
i = 2
fig, axs = plt.subplots(1, 3, figsize=(8, 8))
axs[0].imshow(pattern)
axs[0].set_title('Pattern')
axs[1].imshow(X[i])
axs[1].set_title('Generated image')
axs[2].imshow(exps[i])
axs[2].set_title('Explanation')
[4]:
Text(0.5, 1.0, 'Explanation')
1.2. Declaring and training the model#
Declare a simple LeNet variant and its training routine.
[4]:
class FCNN(nn.Module):
""" Basic NN for image classification. """
def __init__(self, imH, imW, cellH, randomState=1):
super(FCNN, self).__init__()
stride = 1
kSize = 5
torch.manual_seed(randomState)
self.conv1 = nn.Conv2d(3, 6, kernel_size=kSize, stride=stride)
self.H1, self.W1 = floor(((imH - kSize) / stride) + 1), floor(((imW - kSize) / stride) + 1)
self.conv2 = nn.Conv2d(6, 3, kernel_size=kSize, stride=stride)
self.H2, self.W2 = floor(((self.H1 - kSize) / stride) + 1), floor(((self.W1 - kSize) / stride) + 1)
self.conv3 = nn.Conv2d(3, 1, kernel_size=kSize, stride=stride)
self.H3, self.W3 = floor(((self.H2 - kSize) / stride) + 1), floor(((self.W2 - kSize) / stride) + 1)
self.fc1 = nn.Linear(self.H3 * self.W3, 100)
self.fc2 = nn.Linear(100, 2)
def forward(self, x):
if len(x.shape) == 3:
# single instance, add the batch dimension
x = x.unsqueeze(0)
x = nn.ReLU()(self.conv1(x))
x = nn.ReLU()(self.conv2(x))
x = nn.ReLU()(self.conv3(x))
x = nn.ReLU()(self.fc1(x.view(x.shape[0], -1)))
x = self.fc2(x)
return x
import copy
# sample training function for classification
def train_net(model, data, criterion, optimizer, device, batchSize, nEpochs, randomState=888):
""" data: dict with 'train' and 'val' entries. Each entry is a list with X: FloatTensor, y: LongTensor """
torch.manual_seed(randomState)
bestValAcc = -np.inf
bestModelWeights = copy.deepcopy(model.state_dict())
for epoch in range(nEpochs):
for phase in ['train', 'val']:
if phase == 'train':
model.train()
else:
model.eval()
lossVal = .0
corrects = 0
for batch in range(int(len(data[phase][0]) / batchSize)):
XBatch = data[phase][0][batch:batch + batchSize].to(device)
yBatch = data[phase][1][batch:batch + batchSize].to(device)
model.zero_grad()
with torch.set_grad_enabled(phase == 'train'):
out = model(XBatch)
loss = criterion(out, yBatch)
if phase == 'train':
loss.backward()
optimizer.step()
_, preds = torch.max(out, 1)
lossVal += loss.item() * XBatch.size(0)
corrects += torch.sum(preds == yBatch.data)
epochLoss = lossVal / len(data[phase][0])
epochAcc = corrects.double() / len(data[phase][0])
print(f'{phase} Loss: {round(epochLoss, 4)} Acc: {round(epochAcc.item(), 4)}')
if phase == 'val' and epochAcc > bestValAcc:
bestValAcc = epochAcc
bestModelWeights = copy.deepcopy(model.state_dict())
model.load_state_dict(bestModelWeights)
return model, bestValAcc
We cast the images to torch.Tensor type and get train, validation and test splits.
[6]:
XTrain, XTest, yTrain, yTest, expsTrain, expsTest = train_test_split(X, y, exps, train_size=0.8, random_state=7)
XTrain, XVal, yTrain, yVal, expsTrain, expsVal = train_test_split(XTrain, yTrain, expsTrain, train_size=0.75, random_state=7)
XTrain = torch.FloatTensor(XTrain).permute(0, 3, 1, 2)
yTrain = torch.LongTensor(yTrain)
XVal = torch.FloatTensor(XVal).permute(0, 3, 1, 2)
yVal = torch.LongTensor(yVal)
XTest = torch.FloatTensor(XTest).permute(0, 3, 1, 2)
yTest = torch.LongTensor(yTest)
[7]:
print(f'Data proportions -> Train: {round(len(XTrain) / len(X), 3)}, Val: {round(len(XVal) / len(X), 3)}, Test: {round(len(XTest) / len(X), 3)}')
print(f'Positive label proportions -> Train: {round((sum(yTrain) / len(yTrain)).item(), 3)}, \
Val: {round((sum(yVal) / len(yVal)).item(), 3)}, Test: {round((sum(yTest) / len(yTest)).item(), 3)}')
Data proportions -> Train: 0.6, Val: 0.2, Test: 0.2
Positive label proportions -> Train: 0.489, Val: 0.526, Test: 0.507
and train the network
[ ]:
nFeatures = len(XTrain[0].flatten())
criterion = nn.CrossEntropyLoss()
model = FCNN(imH=32, imW=32, cellH=4)
optimizer = optim.Adam(model.parameters(), lr=1e-3)
data = {'train': [XTrain, yTrain], 'val': [XVal, yVal]}
if torch.cuda.is_available():
device = torch.device("cuda:0")
model.to(device)
data["train"] = [XTrain.to(device), yTrain.to(device)]
data["val"] = [XVal.to(device), yVal.to(device)]
else:
device = torch.device("cpu")
model, valAcc = train_net(model, data, criterion, optimizer, device, batchSize=10, nEpochs=10, randomState=7)
[9]:
if torch.cuda.is_available():
print(f'Validation F1: {round(f1_score(yVal, F.softmax(model(XVal.cuda()), dim=-1).cpu().argmax(dim=1).numpy()), 3)}')
print(f'Test F1: {round(f1_score(yTest, F.softmax(model(XTest.cuda()), dim=-1).cpu().argmax(dim=1).numpy()), 3)}')
else:
print(f'Validation F1: {round(f1_score(yVal, F.softmax(model(torch.FloatTensor(XVal)), dim=-1).argmax(dim=1).detach().numpy()), 3)}')
print(f'Test F1: {round(f1_score(yTest, F.softmax(model(torch.FloatTensor(XTest)), dim=-1).argmax(dim=1).detach().numpy()), 3)}')
Validation F1: 0.988
Test F1: 0.983
1.3. Generating and evaluating explanations#
With the model trained on the synthetic images, we generate explanations (with Captum, but feel free to use other methods!). First, declare the explainers:
[10]:
layer = [layer for _, layer in model.named_modules()][-3]
gradShap = GradientShap(model)
intGrad = IntegratedGradients(model)
occlusion = Occlusion(model)
deepLift = DeepLift(model)
guidedBackProp = GuidedBackprop(model)
guidedGradCAM = GuidedGradCam(model, layer)
And define a function to obtain the explanations from different methods:
[5]:
def get_attributions(data, targets, explainer, params=None):
"""
:param data: (Tensor) data to explain
:param targets: (Tensor) class labels w.r.t which we want to compute the attributions
:param explainer: (captum.attr method) initialised explainer
:param params: (dict) parameters for the .attribute method of the explainer
:return: ndarray of shape with attributions
"""
if params is None:
params = {}
elif "baselines" in params and type(params["baselines"]) != int:
params["baselines"] = params["baselines"].to(device)
attributions = []
for image, target in zip(data, targets):
attr = explainer.attribute(image.unsqueeze(0), target=target, **params).cpu().squeeze().detach().numpy()
# mean pool channel attributions
attr = np.mean(attr, axis=0)
# viz._normalize_image_attr(tmp, 'absolute_value', 10)
attributions.append(_minmax_normalize_array(attr))
return np.array(attributions)
[ ]:
# use predicted labels
obsToExplain = torch.FloatTensor(XTest[:5]).to(device)
expsToCompare = expsTest[:5]
predTargets = F.softmax(model(obsToExplain), dim=-1).argmax(dim=1)[:5].to(device)
z = torch.LongTensor([1 if e == 0 else 0 for e in predTargets])
# z = torch.zeros(len(predTargets), dtype=torch.int)
[ ]:
# takes some minutes to run
gradShapExpsTest = get_attributions(obsToExplain, predTargets, gradShap, {'baselines': torch.zeros((1, 3, imH, imW))})
intGradExpsTest = get_attributions(obsToExplain, predTargets, intGrad)
deepLiftExpsTest = get_attributions(obsToExplain, predTargets, deepLift)
occlusionExpsTest = get_attributions(obsToExplain, predTargets, occlusion, {'baselines': 0, 'sliding_window_shapes': (3, cellH*2, cellW*2)})
gBackPropExpsTest = get_attributions(obsToExplain, z, guidedBackProp)
gGradCAMExpsTest = get_attributions(obsToExplain, z, guidedGradCAM)
with open('expsSynth.pickle', 'wb') as handle:
pickle.dump([gradShapExpsTest, intGradExpsTest, deepLiftExpsTest, occlusionExpsTest, gBackPropExpsTest, gGradCAMExpsTest], handle)
[ ]:
with open('expsSynth.pickle', 'rb') as handle:
gradShapExpsTest, intGradExpsTest, deepLiftExpsTest, occlusionExpsTest,\
gBackPropExpsTest, gGradCAMExpsTest = pickle.load(handle)
[ ]:
i = 3
fig, axs = plt.subplots(1, 8, figsize=(15,15))
axs[0].imshow(obsToExplain[i].cpu().permute(1, 2, 0))
axs[0].set_title('Image')
axs[1].imshow(expsToCompare[i])
axs[1].set_title('g.t. explanation')
axs[2].imshow(intGradExpsTest[i])
axs[2].set_title('integratedGradient')
axs[3].imshow(gradShapExpsTest[i])
axs[3].set_title('gradientSHAP')
axs[4].imshow(deepLiftExpsTest[i])
axs[4].set_title('deepLift')
axs[5].imshow(occlusionExpsTest[i])
axs[5].set_title('occlusion')
axs[6].imshow(gBackPropExpsTest[i])
axs[6].set_title('Guided Backprop.')
axs[7].imshow(gGradCAMExpsTest[i])
axs[7].set_title('Guided gradCAM.')
Text(0.5, 1.0, 'Guided gradCAM.')
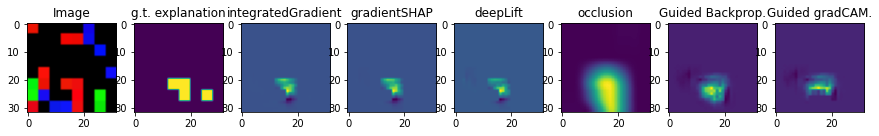
And we can evaluate the explanations. We set the binarization threshold for the generated explanations to 0.5 because we only want high values to count.
[ ]:
metrics = ['auc', 'fscore', 'prec', 'rec', 'cs']
gradShapScores = saliency_map_scores(expsTest[yTest == 1], gradShapExpsTest, metrics=metrics, binThreshold=0.5)
intGradScores = saliency_map_scores(expsTest[yTest == 1], intGradExpsTest, metrics=metrics, binThreshold=0.5)
deepLiftScores = saliency_map_scores(expsTest[yTest == 1], deepLiftExpsTest, metrics=metrics, binThreshold=0.5)
occlusionScores = saliency_map_scores(expsTest[yTest == 1], occlusionExpsTest, metrics=metrics, binThreshold=0.5)
gBackPropScores = saliency_map_scores(expsTest[yTest == 1], gBackPropExpsTest, metrics=metrics, binThreshold=0.5)
gGradCAMScores = saliency_map_scores(expsTest[yTest == 1], gGradCAMExpsTest, metrics=metrics, binThreshold=0.5)
scores = pd.DataFrame(data=[gradShapScores, intGradScores, deepLiftScores,
occlusionScores, gBackPropScores, gGradCAMScores], columns=metrics)
scores['technique'] = ['gradSHAP', 'intGrad', 'deepLift', 'occlusion', 'guidedBackProp', 'guidedGradCAM']
scores
Note how the warnings tell us that the predictions from one of the techniques did not contain any relevant values.
From here, we can build a more complex explanation evaluation pipeline. Suppose that, given some explainer architecture and model, we want to measure how the influence of some explainer hyperparameters influence the quality of their generated explanations.
[6]:
def eval_explainers(
explainers,
explainerConfigs,
data,
targets,
trueExps,
metrics,
binThreshold: float = .5) -> dict:
"""
:param explainers: (dict of captum.attr explainers) explainers to use
:param explainerConfigs: (dict of list of dict) hyperparameter values to use for each explainer (see Captum docs)
:param data: (Tensor) data to explain
:param targets: (Tensor) labels w.r.t. which we compute the explanations
:param trueExps: (ndarray) ground truth explanations
:param metrics: (array-like of str) metrics to compute
:param float binThreshold: threshold to use when binarizing for the computation of classification metrics.
"""
allScores = {explainer: {met: [] for met in metrics} for explainer in explainers.keys()}
for explainerName, explainer in explainers.items():
for config in explainerConfigs[explainerName]:
exps = get_attributions(data, targets, explainer, config)
evals = saliency_map_scores(trueExps, exps, metrics=metrics, binThreshold=binThreshold)
for i, score in enumerate(evals):
allScores[explainerName][metrics[i]].append(score)
return allScores
for example, given these gradSHAP and guidedGradCAM configurations:
[ ]:
configs = {
'gradSHAP': [{'baselines': torch.zeros((1, 3, imH, imW)), 'n_samples': 10, 'stdevs': 0.1},
{'baselines': torch.zeros((1, 3, imH, imW)), 'n_samples': 15, 'stdevs': 0.15}],
'gradCAM': [{'interpolate_mode': 'nearest'},
{'interpolate_mode': 'area'}]
}
explainers = {'gradSHAP': gradShap, 'gradCAM': guidedGradCAM}
metrics = ['auc', 'fscore', 'prec', 'rec', 'cs']
[ ]:
# takes some minutes to run
scores = eval_explainers(explainers, configs, XTest[yTest==1].to(device), yTest[yTest==1], expsTest[yTest==1], metrics)
with open('synthScores.pickle', 'wb') as f:
pickle.dump(scores, f)
[ ]:
with open('synthScores.pickle', 'rb') as f:
scores = pickle.load(f)
For each explainer and configuration, we have a score:
[ ]:
scores
{'gradSHAP': {'auc': [0.7992577, 0.80201876],
'fscore': [0.5050217, 0.5090714],
'prec': [0.9954608, 0.9955106],
'rec': [0.34479782, 0.3487118],
'cs': [0.39023715, 0.3968119]},
'gradCAM': {'auc': [0.34990147, 0.31682178],
'fscore': [0.1094004, 0.11006599],
'prec': [0.05815248, 0.058509283],
'rec': [0.9214127, 0.92628205],
'cs': [0.23264565, 0.23132235]}}
With these metrics, then, we can evaluate the performance of explainers.
2. Evaluating Kahikatea image explanations#
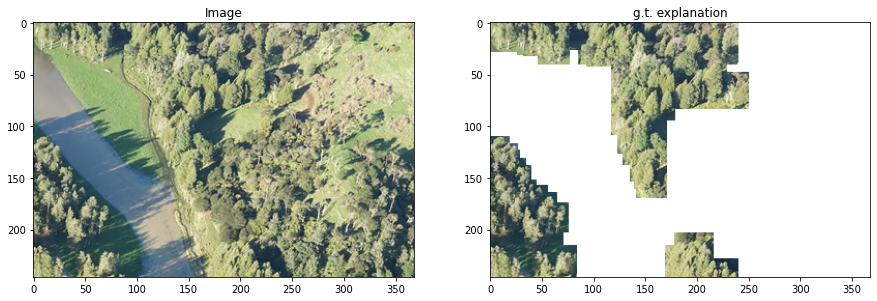
teex includes real datasets with available ground truth explanations. For example, the Kahikatea dataset contains 519 images, and the task is to tell whether each observation contains Kahikatea trees or not. There are 232 positive observations and 287 negative ones.
In teex, the non-artificial datasets are implemented as classes, similarly to PyTorch. After instancing the class, the data itself will be downloaded if it has not been used before. Once done, one can slice it to obtain observations. Each observation contains the data point, the label and the ground truth explanation.
[7]:
from teex.saliencyMap.data import Kahikatea
[8]:
nClasses = 2
kahikateaData = Kahikatea()
kData, kLabels, kExps = kahikateaData[:]
Files do not exist or are corrupted:
Downloading https://zenodo.org/record/5059769/files/kahikatea.zip?download=1 to /opt/conda/lib/python3.7/site-packages/teex/_datasets/saliencyMap/kahikatea/rawKahikatea.zip
142303232it [00:43, 3241716.65it/s]
[9]:
i = 0
fig, axs = plt.subplots(1, 2, figsize=(15,15))
axs[0].imshow(kData[i])
axs[0].set_title('Image')
axs[1].imshow(kExps[i])
axs[1].set_title('g.t. explanation')
[9]:
Text(0.5, 1.0, 'g.t. explanation')
[10]:
kahikateaData.classMap
[10]:
{0: 'Not in image', 1: 'In image'}
Let’s fine-tune a pre-trained squeezenet for our particular task.
[34]:
torch.hub._validate_not_a_forked_repo=lambda a,b,c: True #
sqznet = torch.hub.load('pytorch/vision:v0.9.0', 'squeezenet1_0', pretrained=True, progress=False)
torch.manual_seed(7)
Using cache found in /root/.cache/torch/hub/pytorch_vision_v0.9.0
[34]:
<torch._C.Generator at 0x7fd90d5742b0>
And modify its architecture so the shape of the output conforms to our 2-class problem instead of the 1000-class ImageNet.
[35]:
sqznet.classifier[1] = nn.Conv2d(512, nClasses, kernel_size=(1,1), stride=(1,1))
sqznet.num_classes = nClasses
inputSize = 224
Define the required transform for the input images
[13]:
from torchvision import transforms
inputTransform = transforms.Compose([transforms.Resize((inputSize, inputSize)),
transforms.ToTensor(),
transforms.Normalize([0.485, 0.456, 0.406], [0.229, 0.224, 0.225])])
resizeTransform = transforms.Compose([transforms.Resize((inputSize, inputSize)),
transforms.ToTensor()])
Transform the data to torch tensors and create the splits:
[14]:
kData = torch.stack([inputTransform(img) for img in kData])
kExps = torch.stack([resizeTransform(img) if isinstance(img, PIL.Image.Image) else torch.zeros((3, inputSize, inputSize)) for img in kExps]).permute(0, 2, 3, 1)
kExps = kExps.numpy().astype(np.float32) # we need g.t. explanations to be numpy arrays for the evaluation
kLabels = torch.LongTensor(kLabels)
kTrain, kTest, kTrainLabels, kTestLabels, kExpsTrain, kExpsTest = train_test_split(kData, kLabels, kExps, train_size=0.8, random_state=7)
kTrain, kVal, kTrainLabels, kValLabels, kExpsTrain, kExpsVal = train_test_split(kTrain, kTrainLabels, kExpsTrain, train_size=0.75, random_state=7)
[15]:
print(f'Data proportions -> Train: {round(len(kTrain) / len(kData), 3)}, Val: {round(len(kVal) / len(kData), 3)}, Test: {round(len(kTest) / len(kData), 3)}')
print(f'Positive label proportions -> Train: {round((sum(kTrainLabels) / len(kTrainLabels)).item(), 3)}, \
Val: {round((sum(kValLabels) / len(kValLabels)).item(), 3)}, Test: {round((sum(kTestLabels) / len(kTestLabels)).item(), 3)}')
Data proportions -> Train: 0.599, Val: 0.2, Test: 0.2
Positive label proportions -> Train: 0.444, Val: 0.519, Test: 0.385
[ ]:
opt = optim.SGD(sqznet.parameters(), lr=1e-3)
crit = nn.CrossEntropyLoss()
device = torch.device("cuda:0" if torch.cuda.is_available() else "cpu")
batchSize = 10
nEpochs = 25
data = {'train': [kTrain.to(device), kTrainLabels.to(device)], 'val': [kVal.to(device), kValLabels.to(device)]}
sqznet.to(device)
sqznet, valAcc = train_net(sqznet, data, crit, opt, device, batchSize, nEpochs, randomState=7)
with open('sqznetTrained.pickle', 'wb') as f:
pickle.dump(sqznet.state_dict(), f)
[38]:
with open('sqznetTrained.pickle', 'rb') as f:
sqznet.load_state_dict(pickle.load(f))
[37]:
print(f'Validation F1: {round(f1_score(kValLabels, F.softmax(sqznet(kVal.to(device)), dim=-1).cpu().argmax(dim=1).detach().numpy()), 3)}')
print(f'Test F1: {round(f1_score(kTestLabels, F.softmax(sqznet(kTest.to(device)), dim=-1).cpu().argmax(dim=1).detach().numpy()), 3)}')
Validation F1: 0.816
Test F1: 0.643
Let us get some explanations as samples
[77]:
back_hook = 'register_full_backward_hook' if torch.__version__ >= '1.8.0' else 'register_backward_hook'
gradShap = GradientShap(sqznet)
intGrad = IntegratedGradients(sqznet)
occlusion = Occlusion(sqznet)
deepLift = DeepLift(sqznet)
guidedBackProp = GuidedBackprop(sqznet)
guidedGradCAM = GuidedGradCam(sqznet, sqznet.features[12])
# use predicted labels
obsToExplain = torch.FloatTensor(kTest[kTestLabels == 1][:2]).to(device)
expsToCompare = kExpsTest[kTestLabels == 1][:2]
predTargets = F.softmax(sqznet(obsToExplain), dim=-1).argmax(dim=1)
# takes some minutes to run
gradShapExpsTest = get_attributions(obsToExplain, predTargets, gradShap, {'baselines': torch.zeros((1, 3, inputSize, inputSize))})
intGradExpsTest = get_attributions(obsToExplain, predTargets, intGrad)
deepLiftExpsTest = get_attributions(obsToExplain, predTargets, deepLift)
occlusionExpsTest = get_attributions(obsToExplain, predTargets, occlusion, {'baselines': 0, 'sliding_window_shapes': (3, inputSize, round(inputSize))})
gBackPropExpsTest = get_attributions(obsToExplain, predTargets, guidedBackProp)
gGradCAMExpsTest = get_attributions(obsToExplain, predTargets, guidedGradCAM)
/opt/conda/lib/python3.7/site-packages/captum/attr/_core/guided_backprop_deconvnet.py:65: UserWarning: Setting backward hooks on ReLU activations.The hooks will be removed after the attribution is finished
"Setting backward hooks on ReLU activations."
[43]:
# we binarize the results for easier visualization (this step is implicitly done by teex)
# when computing metrics.
from teex._utils._arrays import _binarize_arrays
gsh = _binarize_arrays(gradShapExpsTest, method='abs', threshold=0.55)
ig = _binarize_arrays(intGradExpsTest, method='abs', threshold=0.55)
dl = _binarize_arrays(deepLiftExpsTest, method='abs', threshold=0.55)
oc = _binarize_arrays(occlusionExpsTest, method='abs', threshold=0.55)
gbp = _binarize_arrays(gBackPropExpsTest, method='abs', threshold=0.55)
ggc = _binarize_arrays(gGradCAMExpsTest, method='abs', threshold=0.55)
i = 0
fig, axs = plt.subplots(1, 8, figsize=(15,15))
axs[0].imshow(obsToExplain[i].cpu().permute(1, 2, 0))
axs[0].set_title('Image')
axs[1].imshow(expsToCompare[i])
axs[1].set_title('g.t. explanation')
axs[2].imshow(ig[i])
axs[2].set_title('integratedGradient')
axs[3].imshow(gsh[i])
axs[3].set_title('gradientSHAP')
axs[4].imshow(dl[i])
axs[4].set_title('deepLift')
axs[5].imshow(oc[i])
axs[5].set_title('occlusion')
axs[6].imshow(gbp[i])
axs[6].set_title('Guided Backprop.')
axs[7].imshow(ggc[i])
axs[7].set_title('Guided gradCAM.')
[43]:
Text(0.5, 1.0, 'Guided gradCAM.')
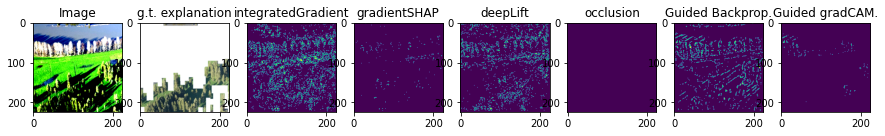
From this visualization, it is clear that the threshold level is an important hyperparameter to consider.
Now, let’s evaluate the quality of the explanations:
[66]:
kExplainers = {'gradSHAP': GradientShap(sqznet),
'gradCAM': GuidedGradCam(sqznet, sqznet.features[12]),
'deepLift': DeepLift(sqznet),
'backProp': GuidedBackprop(sqznet),
'occlusion': Occlusion(sqznet),
'intGrad': IntegratedGradients(sqznet)}
kConfigs = {
'gradSHAP': [{'baselines': torch.zeros((1, 3, inputSize, inputSize)), 'n_samples': 5, 'stdevs': 0.1}],
'gradCAM': [{'interpolate_mode': 'nearest'}],
'deepLift': [{}],
'backProp': [{}],
'occlusion': [{'baselines': 0, 'sliding_window_shapes': (3, round(inputSize), round(inputSize))}],
'intGrad': [{'method': 'riemann_trapezoid'}]
}
metrics = ['auc', 'fscore', 'prec', 'rec', 'cs']
binThresholds = [e / 100 for e in range(10, 70, 5)]
Evaluate positive test explanations. Note that teex implicitly handles the conversion of the RGB masks into 0-1 normalised grayscale masks (the shape of the g.t.s need to be (imH, imW, 3) for it to happen).
[67]:
# use predicted labels
obsToExplain = torch.FloatTensor(kTest[kTestLabels == 1]).to(device)
expsToCompare = kExpsTest[kTestLabels == 1]
predTargets = F.softmax(sqznet(obsToExplain), dim=-1).argmax(dim=1)
[ ]:
# comparison to ground truth explanations
resGT = {}
for binThres in binThresholds:
# takes a few minutes to run
scoresK = eval_explainers(kExplainers, kConfigs, obsToExplain, predTargets,
expsToCompare, metrics, binThreshold=binThres)
resGT[f"{binThres}"] = scoresK
# fileName = f'kahikateaScoresThres{binThres}.pickle'
# with open(fileName, 'wb') as f:
# pickle.dump(scoresK, f)
with open("kahikateaScores", 'wb') as f:
pickle.dump(resGT, f)
[ ]:
resGT
[ ]:
# comparison to integrated gradients explanations
obsToExplain = torch.FloatTensor(kTest[kTestLabels == 1]).to(device)
predTargets = F.softmax(sqznet(obsToExplain), dim=-1).argmax(dim=1)
expsToCompare = get_attributions(obsToExplain, predTargets, intGrad)
resIntGrad = {}
for binThres in binThresholds:
# takes a few minutes to run
scoresK = eval_explainers(kExplainers, kConfigs, obsToExplain, predTargets,
expsToCompare, metrics, binThreshold=binThres)
resIntGrad[f"{binThres}"] = scoresK
with open("kahikateaScores", 'wb') as f:
pickle.dump(resIntGrad, f)
[ ]:
resIntGrad